About Bio-jETI
Bio-jETI is a service platform for interdisciplinary work on biological application domains. It uses the jETI service integration technology for remote tool integration and the jABC framework as a graphical workflow modeling tool. Bio-jETI, domain experts, like biologists who are not trained in computer science, can directly define complex service orchestrations as workflow models and use efficient and complex bioinformatics tools in a simple and intuitive way.
Workflow management
The jETI technology makes it possible to combine heterogeneous remote services, such as bioinformatics analysis tools and information repositories, in a seamless way. Tools of different kinds (e.g. stand-alone and legacy tools, but also REST and web services), from different providers, and also from different application domains can be managed by the jETI toolservers, that provide homogeneous interfaces to their client applications, for instance the jABC.
The jABC is a framework for graphical workflow coordination that fully implements the concepts of service oriented computing. The services provided via jETI constitute libraries of basic services that can then be orchestrated within the jABC. With this framework, users are able to assemble and manage their analysis workflows in a graphical and intuitive way.
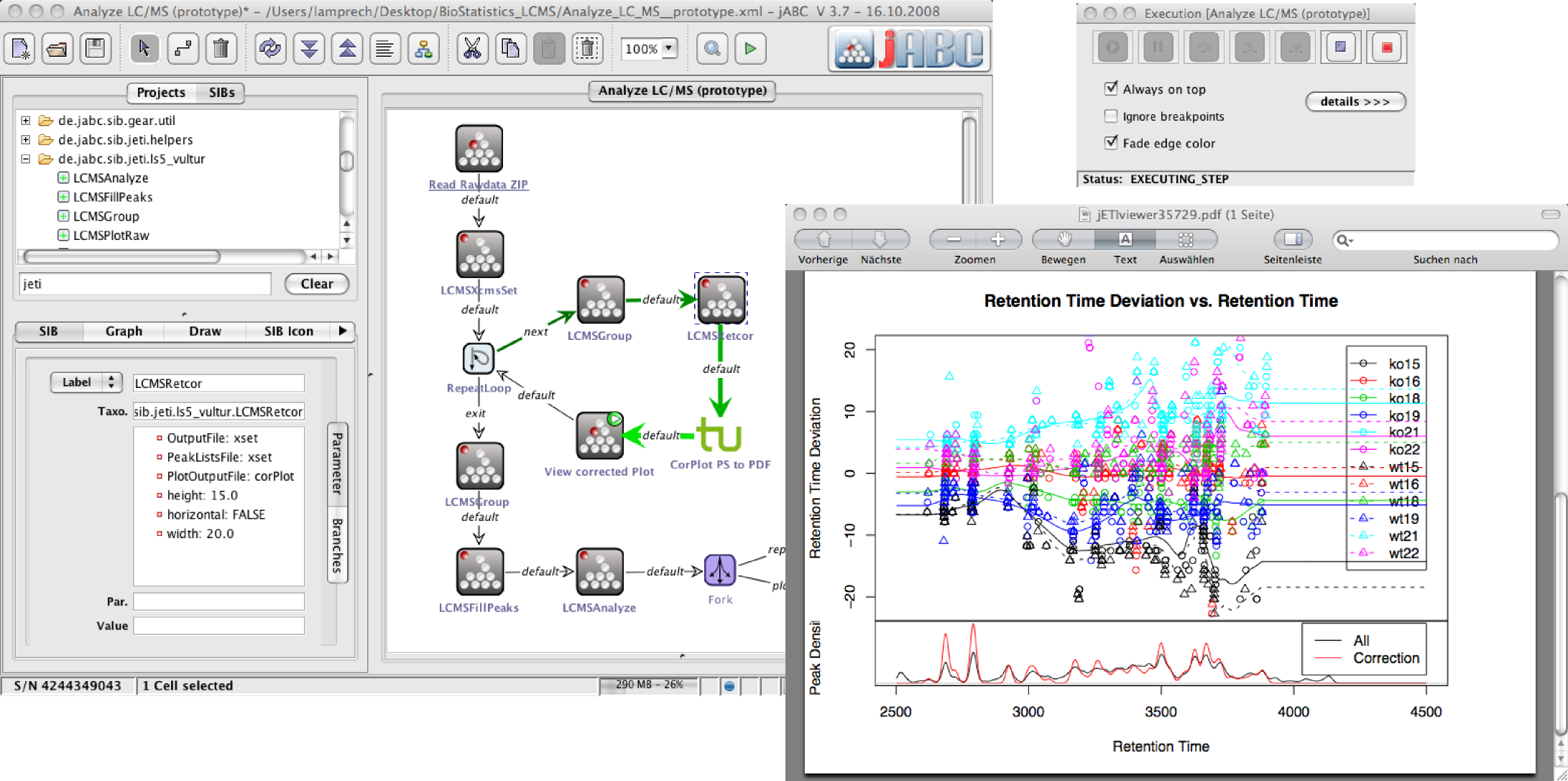 The picture at the right shows the Bio-jETI user interface during workflow execution. The depicted workflow performs the analysis of LC/MS data and resulted from a joint project with the Center for Applied Proteomics (ZAP) Dortmund. The input data was provided by the participating biologists, while the statisticians developed a set of R scripts for constituent analysis steps. With Bio-jETI we integrated the scripts and further data management building blocks, and coordinated the tools and the data in such a way as to enable them to define the whole analysis workflow.
The picture at the right shows the Bio-jETI user interface during workflow execution. The depicted workflow performs the analysis of LC/MS data and resulted from a joint project with the Center for Applied Proteomics (ZAP) Dortmund. The input data was provided by the participating biologists, while the statisticians developed a set of R scripts for constituent analysis steps. With Bio-jETI we integrated the scripts and further data management building blocks, and coordinated the tools and the data in such a way as to enable them to define the whole analysis workflow.
(Semi-) Automatic Worfklow Design
The PROPHETS plugin extends Bio-jETI by functionality for semantics-based, (semi-) automatic worfklow design. With this plugin, users can use workflow synthesis methodology to create parts of the workflow models automatically according to abstract specifications.
Methods for the automatic composition of services into executable workflows need detailed knowledge about the application domain, in particular about the available services and their behavior in terms of input/output data descriptions. Most Bio-jETI domain models are based on the EMBRACE Data and Methods Ontology (EDAM), which provides a comprehensive controlled vocabulary for the annotation of bioinformatics resources.
Features
Bio-jETI's features at a glance:- Easy integration of services via the jETI technology.
- Classification of services using domain-specific, adaptable taxonomies.
- Graphical workflow definition.
- Hierarchical models, support of subworkflows.
- Direct execution of the models.
- Generation of stand-alone source code for independent and repeated execution of a workflow.
- Formal verification of models (embedded modelchecker).
- Semantics-based, (semi-) automatic worfklow design.